This is not a proper blog post yet, just my notes.
pandas (TODO)1. What is pandas and why would you use it?
pandasis a Python package created for working with tables known asDataFrames;it is the only reasonable Python package for this purpose, which makes Python a little modest comparing to R (base, data.table, dplyr - every one of them has a better interface than pandas) when we process tables;
Even though I use pandas almost every day, there are certain solutions that I constantly forget about.
2. Examples
One of the simplest, yet powerful interfaces to work with tables has SQL, so I will describe pandas’ equivalents to SQL’s functions.
But first let’s prepare a dataset:
import pandas as pd
import re
from sklearn.datasets import load_iris
import matplotlib.pyplot as plt
import numpy as np
def prepare_iris_as_in_r():
iris_raw = load_iris()
colnames = [re.sub(' ', '_', re.sub(' \(cm\)', '', x))
for x in iris_raw.feature_names]
iris = pd.DataFrame(iris_raw.data, columns=colnames)
species = pd.DataFrame({'species_index': range(3),
'species': iris_raw.target_names})
iris['species_index'] = iris_raw.target
iris = pd.merge(iris, species, on='species_index')
iris.drop('species_index', axis=1, inplace=True)
return irisWe’ve been working on this dataset in almost every post I wrote for this blog. It’s rather small and simple, so we will not take advantage of pandas’s processing efficiency.
iris = prepare_iris_as_in_r()reading data from a file
In our examples we will not be doing this, because we have already loeaded the data from sklearn. You can do it with:
data = pd.read_csv()Don’t name your datasets
data. Everything we work on is data, so this name conveys no information. I named our dataset that way for the same reason as in most tutorials the default password is “password”.
useful parameters:
sepdelimiterheader
Parameters are rather self-explanatory. And yes, I know that in SQL we don’t read files ;)
WHERE filtering
The easiest way to filter a DataFrame is by using:
iris[iris.species == "setosa"].head()
iris[~iris.sepal_length.isna()].head()SELECT 2 selecting
iris[['species', 'sepal_width']]## species sepal_width
## 0 setosa 3.5
## 1 setosa 3.0
## 2 setosa 3.2
## 3 setosa 3.1
## 4 setosa 3.6
## 5 setosa 3.9
## 6 setosa 3.4
## 7 setosa 3.4
## 8 setosa 2.9
## 9 setosa 3.1
## 10 setosa 3.7
## 11 setosa 3.4
## 12 setosa 3.0
## 13 setosa 3.0
## 14 setosa 4.0
## 15 setosa 4.4
## 16 setosa 3.9
## 17 setosa 3.5
## 18 setosa 3.8
## 19 setosa 3.8
## 20 setosa 3.4
## 21 setosa 3.7
## 22 setosa 3.6
## 23 setosa 3.3
## 24 setosa 3.4
## 25 setosa 3.0
## 26 setosa 3.4
## 27 setosa 3.5
## 28 setosa 3.4
## 29 setosa 3.2
## .. ... ...
## 120 virginica 3.2
## 121 virginica 2.8
## 122 virginica 2.8
## 123 virginica 2.7
## 124 virginica 3.3
## 125 virginica 3.2
## 126 virginica 2.8
## 127 virginica 3.0
## 128 virginica 2.8
## 129 virginica 3.0
## 130 virginica 2.8
## 131 virginica 3.8
## 132 virginica 2.8
## 133 virginica 2.8
## 134 virginica 2.6
## 135 virginica 3.0
## 136 virginica 3.4
## 137 virginica 3.1
## 138 virginica 3.0
## 139 virginica 3.1
## 140 virginica 3.1
## 141 virginica 3.1
## 142 virginica 2.7
## 143 virginica 3.2
## 144 virginica 3.3
## 145 virginica 3.0
## 146 virginica 2.5
## 147 virginica 3.0
## 148 virginica 3.4
## 149 virginica 3.0
##
## [150 rows x 2 columns]proper ways of selection + filtering
loc - you may use ranges as well as names
iris.loc[:10, ['species', 'sepal_length']]## species sepal_length
## 0 setosa 5.1
## 1 setosa 4.9
## 2 setosa 4.7
## 3 setosa 4.6
## 4 setosa 5.0
## 5 setosa 5.4
## 6 setosa 4.6
## 7 setosa 5.0
## 8 setosa 4.4
## 9 setosa 4.9
## 10 setosa 5.4iloc - you can use unly ranges
iris.iloc[:10, :3]## sepal_length sepal_width petal_length
## 0 5.1 3.5 1.4
## 1 4.9 3.0 1.4
## 2 4.7 3.2 1.3
## 3 4.6 3.1 1.5
## 4 5.0 3.6 1.4
## 5 5.4 3.9 1.7
## 6 4.6 3.4 1.4
## 7 5.0 3.4 1.5
## 8 4.4 2.9 1.4
## 9 4.9 3.1 1.5GROUP BY 3 aggregating
iris.groupby('species').agg({'sepal_length': sum, 'petal_length': np.median})## petal_length sepal_length
## species
## setosa 1.50 250.3
## versicolor 4.35 296.8
## virginica 5.55 329.4pd.DataFrame({'count' : df1.groupby( [ "Name", "City"] ).size()}).reset_index()JOIN 4 joining
plotting
iris.groupby('species').size().plot.bar()
plt.show()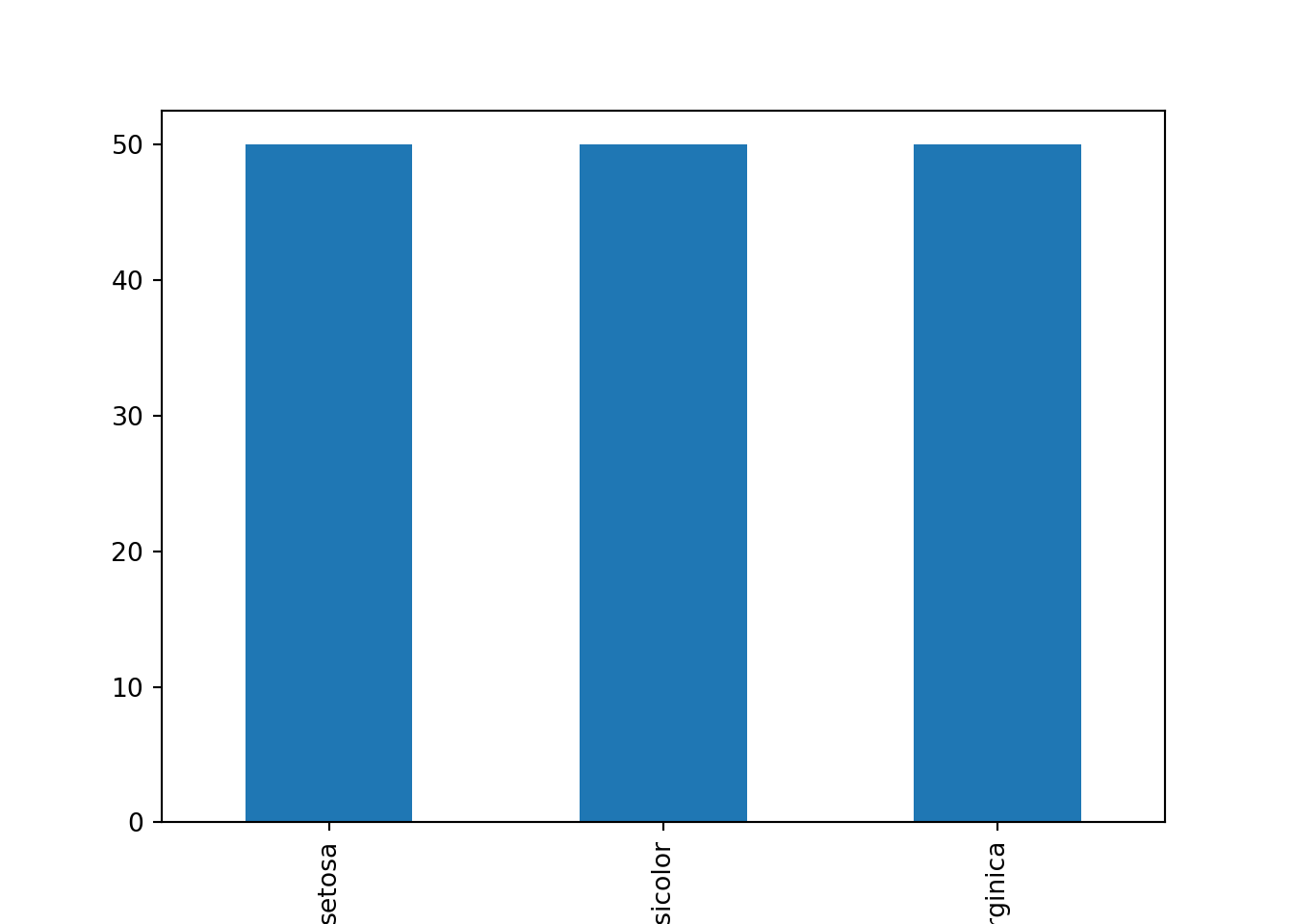
ordering
pivot table
indexes, or Series vs. DataFrame
You may have noticed that pandas uses indexes extensively, which may be not very intuitive if you come with R or SQL background (especially that index in pandas is means something different than in SQL, which is misleading). In general the easiest way to cope with their problematic nature is trying to avoid them.
plt.scatter(iris.sepal_length, iris.sepal_width)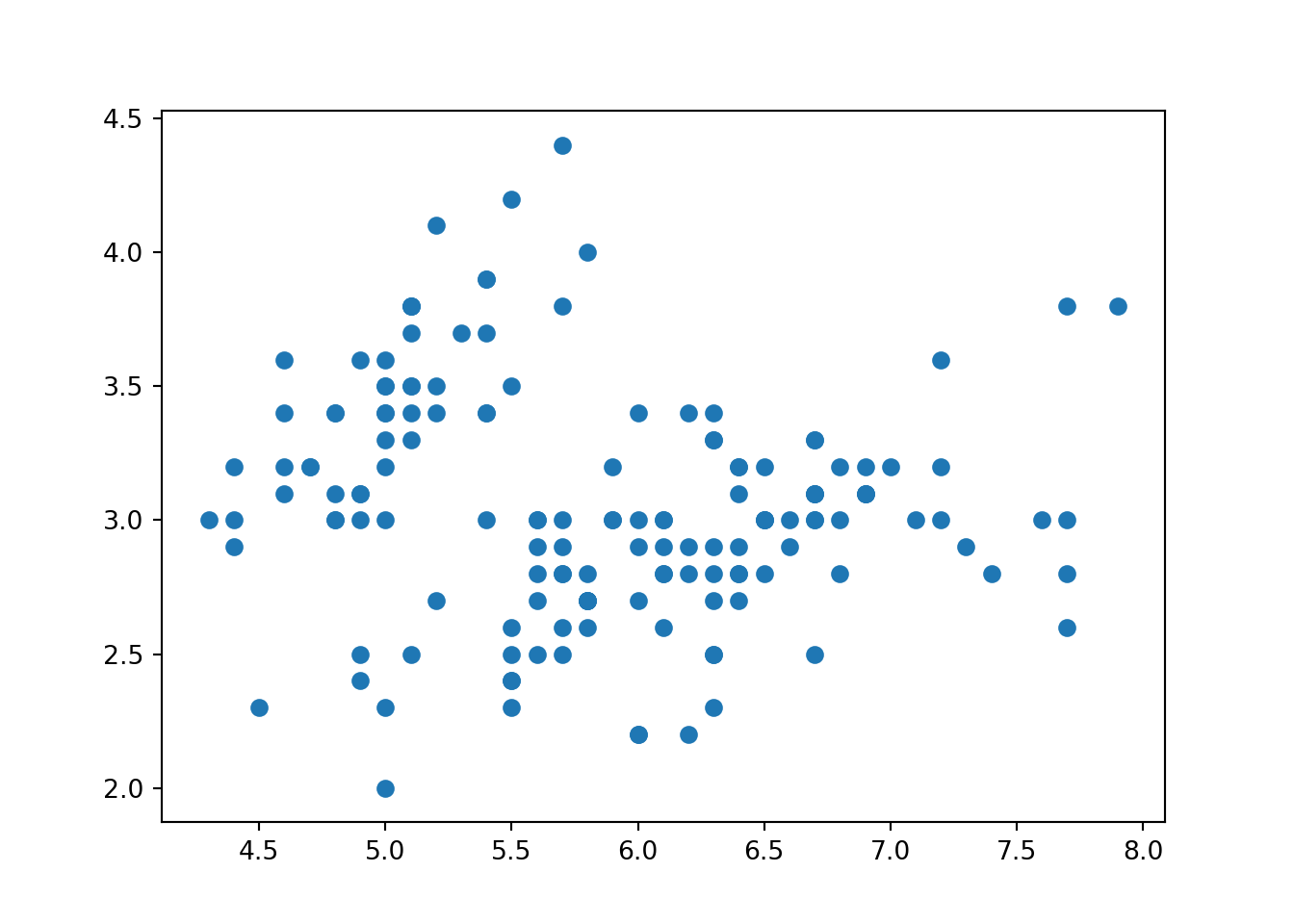
TODO
import numpy as np
col1 = list('abcdefghij')
start_date = pd.date_range(pd.datetime.today(), periods=10).tolist()
end_date = pd.date_range(pd.datetime.today(), periods=10).tolist()
df = pd.DataFrame(dict(col1=col1, start_date=start_date, end_date=end_date))
df.melt(id_vars='col1',
value_vars=['start_date', 'end_date'],
var_name='is_start_end',
value_name='date')## col1 is_start_end date
## 0 a start_date 2019-12-22 22:54:59.241695
## 1 b start_date 2019-12-23 22:54:59.241695
## 2 c start_date 2019-12-24 22:54:59.241695
## 3 d start_date 2019-12-25 22:54:59.241695
## 4 e start_date 2019-12-26 22:54:59.241695
## 5 f start_date 2019-12-27 22:54:59.241695
## 6 g start_date 2019-12-28 22:54:59.241695
## 7 h start_date 2019-12-29 22:54:59.241695
## 8 i start_date 2019-12-30 22:54:59.241695
## 9 j start_date 2019-12-31 22:54:59.241695
## 10 a end_date 2019-12-22 22:54:59.249235
## 11 b end_date 2019-12-23 22:54:59.249235
## 12 c end_date 2019-12-24 22:54:59.249235
## 13 d end_date 2019-12-25 22:54:59.249235
## 14 e end_date 2019-12-26 22:54:59.249235
## 15 f end_date 2019-12-27 22:54:59.249235
## 16 g end_date 2019-12-28 22:54:59.249235
## 17 h end_date 2019-12-29 22:54:59.249235
## 18 i end_date 2019-12-30 22:54:59.249235
## 19 j end_date 2019-12-31 22:54:59.249235